Demo: fMRI searchlight with naive bayes classifier
The data used here is available from http://cosmomvpa.org/datadb.zip
This example uses the following dataset: + 'digit' A participant made finger pressed with the index and middle finger of the right hand during 4 runs in an fMRI study. Each run was divided in 4 blocks with presses of each finger and analyzed with the GLM, resulting in 2*4*4=32 t-values
This example uses the cosmo_naive_bayes_classifier_searchlight, which is a fast alternative to using the regular searchlight with a crossvalidation measure and a classifier
- For CoSMoMVPA's copyright information and license terms, #
- see the COPYING file distributed with CoSMoMVPA. #
Contents
Set data paths
The function cosmo_config() returns a struct containing paths to tutorial data. (Alternatively the paths can be set manually without using cosmo_config.)
config = cosmo_config(); digit_study_path = fullfile(config.tutorial_data_path, 'digit'); readme_fn = fullfile(digit_study_path, 'README'); cosmo_type(readme_fn); output_path = config.output_data_path; % reset citation list cosmo_check_external('-tic');
Overview
--------
fMRI responses to a human participant pressing buttons with the index and middle finger.
Contents
--------
- glm_T_stats_allruns+orig.{BRIK,HEAD}:
t-statistics associated with finger presses.
There are 4 runs, each with 4 blocks.
Each block has two t-statistics, one for each of button presses for the
index and middle finger (in that order).
- epi+orig.{HEAD,BRIK}:
A single EPI image.
- icoXX_mh.YY.asc:
Surface anatomy meshes of two hemispheres with standard topology, generated
using AFNI SUMA's MapIcosahedron. Left and right hemisphere surfaces were
merged in the order left, right; the first half of the nodes and faces refer
to the left hemisphere, the second half to the right hemisphere.
XX={16,64} is the number of linear divisions of MapIcosahedron; surfaces
have (XX^2)*10+2 nodes and XX^2*20 faces in each hemisphere.
YY={pial_al,white_al} are the outer and inner surfaces around the grey matter
generated by FreeSurfer. YY=intermediate_al is the node-wise average of the
pial and white surfaces; this surface can be used for single-surface
analyses when surfaces are generated using Caret or BrainVoyager
YY=inflated_alCoMmedial are inflated surfaces with the center of mass
along the medial side of the two hemispheres. These surfaces are suitable
for visualization purposes.
Methods
-------
This dataset contains data from a fingerpress experiment where a participant
(28y right-handed male) pressed buttons on a box with the index and middle
finger of the right hand while fMRI volumes were acquired. Over four runs, in
total sixteen blocks of 72s finger presses were acquired; in each block the
participant pressed the index and middle finger during different `mini-blocks'
of 8 s each. Each block was preceded and followed by a 16s rest period.
Functional data was preprocessed in AFNI with despiking, time slice correction,
motion correction, and scaling to percent signal change by dividing the signal
for each volume by the mean of the run. No spatial smoothing or interpolation
was applied to the functional data, except for interpolation during motion
correction. The preprocessed data was analyzed with a general linear model
(GLM) with separate regressors for each finger and each block (and some
regressors of no interest) to obtain t-values for each finger in each block.
License
-------
The contents are made available by Nikolaas N. Oosterhof <nikolaas.oosterhof
|at| unitn.it> under the Creative Commons CC0 1.0 Universal Public Domain
Dedication ("CC0"). See the LICENSE file for details, or visit
http://creativecommons.org/publicdomain/zero/1.0/deed.en.
Contact
-------
Nikolaas N. Oosterhof <nikolaas.oosterhof |at| unitn.it>
LDA classifier searchlight analysis
%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% % This analysis identified brain regions where the categories can be % distinguished using an odd-even partitioning scheme and a Naive Bayes % classifier. data_path = digit_study_path; data_fn = fullfile(data_path, 'glm_T_stats_perblock+orig.HEAD'); mask_fn = fullfile(data_path, 'brain_mask+orig.HEAD'); targets = repmat(1:2, 1, 16)'; % class labels: 1 2 1 2 1 2 1 2 1 2 ... 1 2 chunks = floor(((1:32) - 1) / 8) + 1; % run labels: 1 1 1 1 1 1 1 1 2 2 ... 4 4 ds_per_run = cosmo_fmri_dataset(data_fn, 'mask', mask_fn, ... 'targets', targets, 'chunks', chunks); % print dataset fprintf('Dataset input:\n'); cosmo_disp(ds_per_run); % set parameters for naive bayes searchlight (partitions) in a % measure_args struct. measure_args = struct(); % Set partition scheme. odd_even is fast; for publication-quality analysis % nfold_partitioner is recommended. % Alternatives are: % - cosmo_nfold_partitioner (take-one-chunk-out crossvalidation) % - cosmo_nchoosek_partitioner (take-K-chunks-out " "). measure_args.partitions = cosmo_oddeven_partitioner(ds_per_run); % print measure and arguments fprintf('Searchlight measure arguments:\n'); cosmo_disp(measure_args); % Define a neighborhood with approximately 100 voxels in each searchlight. nvoxels_per_searchlight = 100; nbrhood = cosmo_spherical_neighborhood(ds_per_run, ... 'count', nvoxels_per_searchlight);
Dataset input:
.sa
.labels
{ 'fi_i_R1_B01#0_Tstat'
'fi_m_R1_B01#0_Tstat'
'fi_i_R1_B02#0_Tstat'
:
'fi_m_R4_B03#0_Tstat'
'fi_i_R4_B04#0_Tstat'
'fi_m_R4_B04#0_Tstat' }@32x1
.stats
{ 'Ttest(168)'
'Ttest(168)'
'Ttest(168)'
:
'Ttest(168)'
'Ttest(168)'
'Ttest(168)' }@32x1
.targets
[ 1
2
1
:
2
1
2 ]@32x1
.chunks
[ 1
1
1
:
4
4
4 ]@32x1
.a
.vol
.mat
[ -2.5 0 0 71.2
0 -2.5 0 112
0 0 2.5 43
0 0 0 1 ]
.dim
[ 61 84 41 ]
.xform
'scanner_anat'
.fdim
.labels
{ 'i'
'j'
'k' }
.values
{ [ 1 2 3 ... 59 60 61 ]@1x61
[ 1 2 3 ... 82 83 84 ]@1x84
[ 1 2 3 ... 39 40 41 ]@1x41 }
.samples
[ -0.171 1.92 -2.74 ... -0.0821 1.12 0.121
-0.547 2.95 -0.743 ... -1.51 0.771 -0.555
3.08 3.48 -0.321 ... 2.82 1.13 1.65
: : : : : :
1.93 1.72 -3.27 ... 0 0 0
0.688 1.22 -1.44 ... 0 0 0
0.495 0.601 -0.448 ... 0 0 0 ]@32x62454
.fa
.i
[ 38 34 35 ... 34 35 36 ]@1x62454
.j
[ 7 8 8 ... 49 49 49 ]@1x62454
.k
[ 5 5 5 ... 36 36 36 ]@1x62454
Searchlight measure arguments:
.partitions
.train_indices
{ [ 1 [ 9
2 10
3 11
: :
22 30
23 31
24 ]@16x1 32 ]@16x1 }
.test_indices
{ [ 9 [ 1
10 2
11 3
: :
30 22
31 23
32 ]@16x1 24 ]@16x1 }
+00:00:01 [####################] -00:00:00 mean size 100.0
Run the searchlight
nb_results = cosmo_naive_bayes_classifier_searchlight(ds_per_run, ...
nbrhood, measure_args);
+00:00:01 [####################] -00:00:00
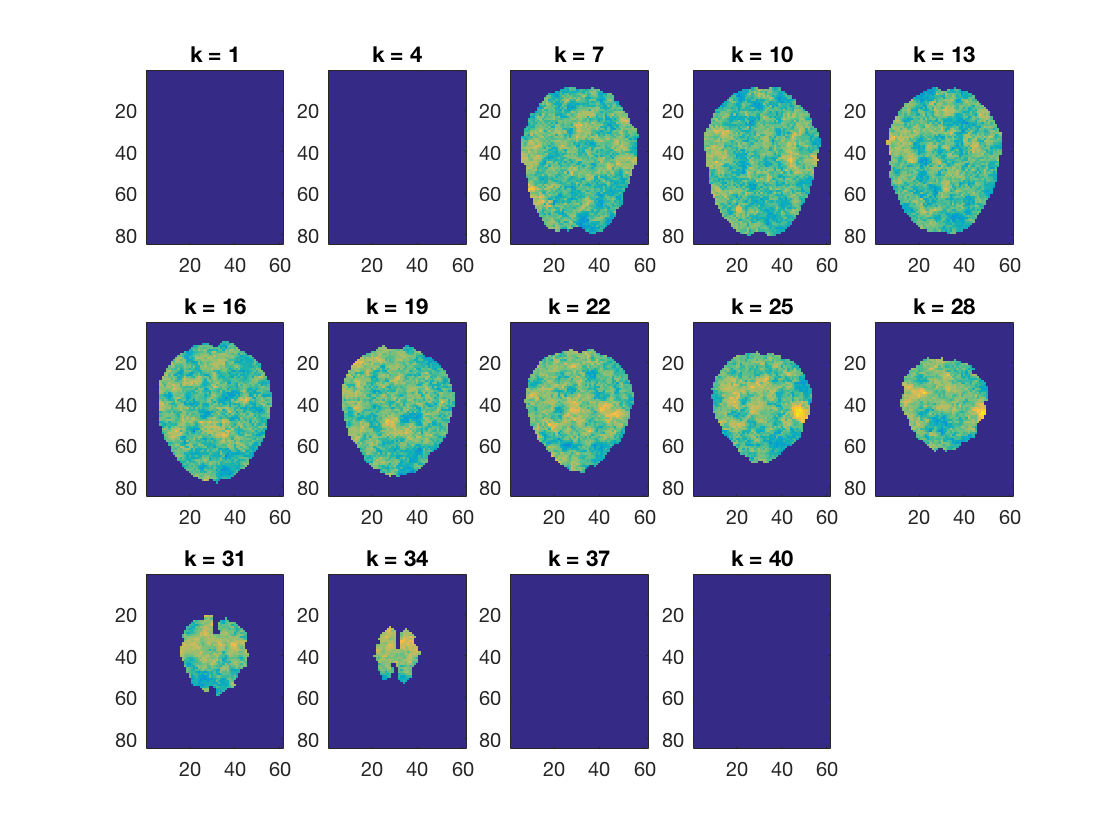
Show results
print output dataset
fprintf('Dataset output:\n'); cosmo_disp(nb_results); % Plot the output cosmo_plot_slices(nb_results); % Define output location output_fn = fullfile(output_path, 'naive_bayes_searchlight+orig'); % Store results to disc cosmo_map2fmri(nb_results, output_fn); % Show citation information cosmo_check_external('-cite');
Dataset output:
.a
.vol
.mat
[ -2.5 0 0 71.2
0 -2.5 0 112
0 0 2.5 43
0 0 0 1 ]
.dim
[ 61 84 41 ]
.xform
'scanner_anat'
.fdim
.labels
{ 'i'
'j'
'k' }
.values
{ [ 1 2 3 ... 59 60 61 ]@1x61
[ 1 2 3 ... 82 83 84 ]@1x84
[ 1 2 3 ... 39 40 41 ]@1x41 }
.fa
.nvoxels
[ 100 104 103 ... 101 99 100 ]@1x62454
.radius
[ 4.12 4.12 3.74 ... 4.12 4.12 4.12 ]@1x62454
.center_ids
[ 1 2 3 ... 6.25e+04 6.25e+04 6.25e+04 ]@1x62454
.i
[ 38 34 35 ... 34 35 36 ]@1x62454
.j
[ 7 8 8 ... 49 49 49 ]@1x62454
.k
[ 5 5 5 ... 36 36 36 ]@1x62454
.samples
[ 0.438 0.438 0.344 ... 0.594 0.594 0.594 ]@1x62454
.sa
.labels
{ 'accuracy' }
If you use CoSMoMVPA and/or some other toolboxes for a publication, please cite:
Z. Saad, G. Chen. AFNI Matlab library. available online from https://github.com/afni/AFNI
N. N. Oosterhof, A. C. Connolly, J. V. Haxby (2016). CoSMoMVPA: multi-modal multivariate pattern analysis of neuroimaging data in Matlab / GNU Octave. Frontiers in Neuroinformatics, doi:10.3389/fninf.2016.00027.. CoSMoMVPA toolbox available online from http://cosmomvpa.org
The Mathworks, Natick, MA, United States. Matlab 24.1.0.2537033 (R2024a) (February 21, 2024). available online from http://www.mathworks.com