Searchlight using a data measure
Using cosmo_searchlight, run cross-validation with nearest neighbor classifier
- For CoSMoMVPA's copyright information and license terms, #
- see the COPYING file distributed with CoSMoMVPA. #
Contents
Define data
config = cosmo_config(); data_path = fullfile(config.tutorial_data_path, 'ak6', 's01'); targets = repmat(1:6, 1, 10); chunks = floor(((1:60) - 1) / 6) + 1; ds = cosmo_fmri_dataset(fullfile(data_path, 'glm_T_stats_perrun.nii'), ... 'mask', fullfile(data_path, 'brain_mask.nii'), ... 'targets', targets, 'chunks', chunks);
Set measure
Use the cosmo_cross_validation_measure and set its parameters (classifier and partitions) in a measure_args struct. >@@>
measure = @cosmo_crossvalidation_measure;
measure_args = struct();
measure_args.classifier = @cosmo_classify_lda;
measure_args.partitions = cosmo_oddeven_partitioner(ds);
% <@@<
Define neighborhood
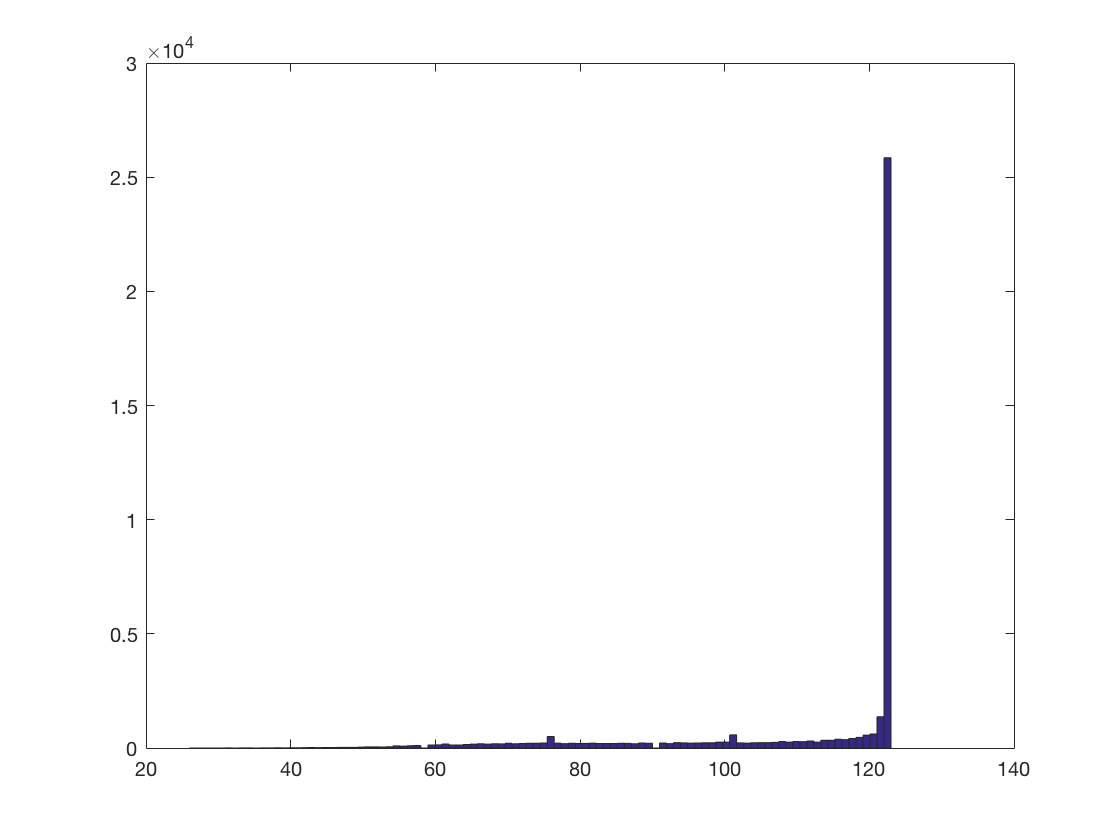
radius = 3; % 3 voxels % define a neighborhood using cosmo_spherical_neighborhood % >@@> nbrhood = cosmo_spherical_neighborhood(ds, 'radius', radius); % <@@< % show a histogram of the number of voxels in each searchlight % >@@> count = cellfun(@numel, nbrhood.neighbors); hist(count, 100); % <@@<
+00:00:00 [####################] -00:00:00 mean size 111.5
Run the searchlight
hint: use cosmo_searchlight with the measure, args and nbrhood
% >@@> results = cosmo_searchlight(ds, nbrhood, measure, measure_args); % <@@< % the following command would store the results to disk: % >> cosmo_map2fmri(results, [data_path 'measure_searchlight.nii']);
+00:00:23 [####################] -00:00:00
Make a histogram of classification accuracies
figure(); hist(results.samples, 47);

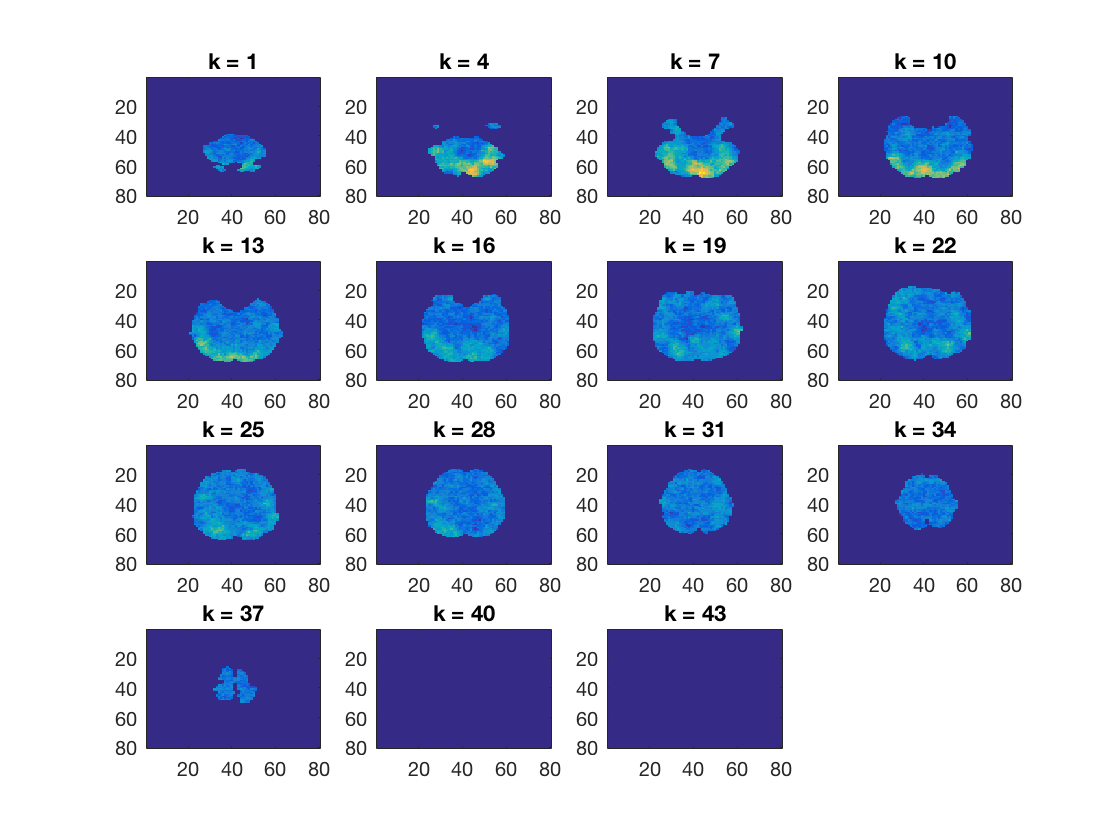
Plot a map
figure(); cosmo_plot_slices(results);