Dataset Basics
Run operations on datasets
- For CoSMoMVPA's copyright information and license terms, #
- see the COPYING file distributed with CoSMoMVPA. #
Contents
Load data and set targets
Load data as before setting targets and chunks appropriately
config = cosmo_config(); data_path = fullfile(config.tutorial_data_path, 'ak6', 's01'); mask_fn = fullfile(data_path, 'vt_mask.nii'); data_fn = fullfile(data_path, 'glm_T_stats_perrun.nii'); ds = cosmo_fmri_dataset(data_fn, 'mask', mask_fn); % set ds.sa.targets (trial conditions) to the 60x1 column vector: % [ 1 2 3 4 5 6 1 2 3 ... 5 6 ]' % >@@> ds.sa.targets = repmat((1:6)', 10, 1); % <@@< % set ds.sa.chunks (acquisition run number) to the 60x1 column vector: % [ 1 1 1 1 1 1 2 2 2 ... 10 10 ]' % >@@> ds.sa.chunks = ceil((1:60)' / 6); % <@@< % set the labels labels = repmat({ 'monkey' 'lemur' 'mallard' 'warbler' 'ladybug' 'lunamoth'}, 10, 1); ds.sa.labels = labels; % This particular ROI has a few constant features due to % not-so-agressive masking, so remove constant features ds = cosmo_remove_useless_data(ds);
Various slicing operations on the samples
% only get data in chunks 1 and 2. % Make a logical mask indicating where .ds.chunks less than or equal to 2 % and store the result in a variable 'chunks12_msk'. % Then use cosmo_slice to select these samples and assign it to a variable % 'ds_chunks12' % (hint: you can use cosmo_match or just '<=') % >@@> chunks12_msk = ds.sa.chunks <= 2; ds_chunks12 = cosmo_slice(ds, chunks12_msk); cosmo_disp(ds_chunks12); % <@@< % As above, but only select data with targets equal to 1 or 3. % (corresponding to {'monkey','mallard'}) % >@@> % (there are multiple ways of doing this) targets_13 = ds.sa.targets == 1 | ds.sa.targets == 3; % element-wise logical 'or' ds_targets13 = cosmo_slice(ds, targets_13); cosmo_disp(ds_targets13); % this is an alternative way to achieve the same goal targets_13_alt = cosmo_match(ds.sa.targets, [1 3]); % using cosmo_match ds_targets13_alt = cosmo_slice(ds, targets_13_alt); cosmo_disp(ds_targets13_alt); % sanity check showing they are the same assert(isequal(ds_targets13, ds_targets13_alt)); % alternative using cosmo_match and the labels labels_monkey_or_mallard = cosmo_match(ds.sa.labels, {'monkey', 'mallard'}); ds_targets13_alt2 = cosmo_slice(ds, labels_monkey_or_mallard); cosmo_disp(ds_targets13_alt2); % sanity check showing they are the same assert(isequal(ds_targets13, ds_targets13_alt2)); % <@@<
.a
.vol
.mat
[ -3 0 0 121
0 3 0 -114
0 0 3 -11.1
0 0 0 1 ]
.xform
'scanner_anat'
.dim
[ 80 80 43 ]
.fdim
.labels
{ 'i'
'j'
'k' }
.values
{ [ 1 2 3 ... 78 79 80 ]@1x80
[ 1 2 3 ... 78 79 80 ]@1x80
[ 1 2 3 ... 41 42 43 ]@1x43 }
.sa
.targets
[ 1
2
3
:
4
5
6 ]@12x1
.chunks
[ 1
1
1
:
2
2
2 ]@12x1
.labels
{ 'monkey'
'lemur'
'mallard'
:
'warbler'
'ladybug'
'lunamoth' }@12x1
.samples
[ 2.73 2.62 2.19 ... 0.669 1.78 2.42
2.51 2.99 3.25 ... 1.06 1.61 0.943
1.3 0.646 0.591 ... 1.51 1.75 3.08
: : : : : :
1.89 1.37 3.05 ... 4.38 4.46 3.86
3.46 3.08 4.2 ... 2.78 1.77 1.19
3.25 2.18 3.99 ... 4.21 4.54 3.41 ]@12x384
.fa
.i
[ 32 33 34 ... 28 29 29 ]@1x384
.j
[ 24 24 24 ... 25 25 26 ]@1x384
.k
[ 3 3 3 ... 9 9 9 ]@1x384
.a
.vol
.mat
[ -3 0 0 121
0 3 0 -114
0 0 3 -11.1
0 0 0 1 ]
.xform
'scanner_anat'
.dim
[ 80 80 43 ]
.fdim
.labels
{ 'i'
'j'
'k' }
.values
{ [ 1 2 3 ... 78 79 80 ]@1x80
[ 1 2 3 ... 78 79 80 ]@1x80
[ 1 2 3 ... 41 42 43 ]@1x43 }
.sa
.targets
[ 1
3
1
:
3
1
3 ]@20x1
.chunks
[ 1
1
2
:
9
10
10 ]@20x1
.labels
{ 'monkey'
'mallard'
'monkey'
:
'mallard'
'monkey'
'mallard' }@20x1
.samples
[ 2.73 2.62 2.19 ... 0.669 1.78 2.42
1.3 0.646 0.591 ... 1.51 1.75 3.08
3.52 2.01 3.32 ... 1.8 2.54 0.949
: : : : : :
2.24 2.37 3.27 ... 4.61 2.13 4.32
2.31 1.61 1.6 ... 0.839 0.544 2.83
2.13 1.22 2.12 ... 2.3 3.14 1.99 ]@20x384
.fa
.i
[ 32 33 34 ... 28 29 29 ]@1x384
.j
[ 24 24 24 ... 25 25 26 ]@1x384
.k
[ 3 3 3 ... 9 9 9 ]@1x384
.a
.vol
.mat
[ -3 0 0 121
0 3 0 -114
0 0 3 -11.1
0 0 0 1 ]
.xform
'scanner_anat'
.dim
[ 80 80 43 ]
.fdim
.labels
{ 'i'
'j'
'k' }
.values
{ [ 1 2 3 ... 78 79 80 ]@1x80
[ 1 2 3 ... 78 79 80 ]@1x80
[ 1 2 3 ... 41 42 43 ]@1x43 }
.sa
.targets
[ 1
3
1
:
3
1
3 ]@20x1
.chunks
[ 1
1
2
:
9
10
10 ]@20x1
.labels
{ 'monkey'
'mallard'
'monkey'
:
'mallard'
'monkey'
'mallard' }@20x1
.samples
[ 2.73 2.62 2.19 ... 0.669 1.78 2.42
1.3 0.646 0.591 ... 1.51 1.75 3.08
3.52 2.01 3.32 ... 1.8 2.54 0.949
: : : : : :
2.24 2.37 3.27 ... 4.61 2.13 4.32
2.31 1.61 1.6 ... 0.839 0.544 2.83
2.13 1.22 2.12 ... 2.3 3.14 1.99 ]@20x384
.fa
.i
[ 32 33 34 ... 28 29 29 ]@1x384
.j
[ 24 24 24 ... 25 25 26 ]@1x384
.k
[ 3 3 3 ... 9 9 9 ]@1x384
.a
.vol
.mat
[ -3 0 0 121
0 3 0 -114
0 0 3 -11.1
0 0 0 1 ]
.xform
'scanner_anat'
.dim
[ 80 80 43 ]
.fdim
.labels
{ 'i'
'j'
'k' }
.values
{ [ 1 2 3 ... 78 79 80 ]@1x80
[ 1 2 3 ... 78 79 80 ]@1x80
[ 1 2 3 ... 41 42 43 ]@1x43 }
.sa
.targets
[ 1
3
1
:
3
1
3 ]@20x1
.chunks
[ 1
1
2
:
9
10
10 ]@20x1
.labels
{ 'monkey'
'mallard'
'monkey'
:
'mallard'
'monkey'
'mallard' }@20x1
.samples
[ 2.73 2.62 2.19 ... 0.669 1.78 2.42
1.3 0.646 0.591 ... 1.51 1.75 3.08
3.52 2.01 3.32 ... 1.8 2.54 0.949
: : : : : :
2.24 2.37 3.27 ... 4.61 2.13 4.32
2.31 1.61 1.6 ... 0.839 0.544 2.83
2.13 1.22 2.12 ... 2.3 3.14 1.99 ]@20x384
.fa
.i
[ 32 33 34 ... 28 29 29 ]@1x384
.j
[ 24 24 24 ... 25 25 26 ]@1x384
.k
[ 3 3 3 ... 9 9 9 ]@1x384
Get sample indices for primates and bugs
Get the sample indices that correspond to primates and bugs, and assign the result to variables 'primate_idx' and 'bug_idx'
% >@@> primate_idx = ds.sa.targets <= 2; bug_idx = ds.sa.targets > 4; % <@@< % Slice the dataset % use the indices in 'primate_idx' and 'bug_idx' as input to cosmo_slice, % and store the result in 'primate_ds' and 'bug_ds', respectively. % >@@> primate_ds = cosmo_slice(ds, primate_idx); bug_ds = cosmo_slice(ds, bug_idx); % <@@<
Subtract mean pattern
Find the mean pattern for primates and bugs and subtract the bug pattern from the primate pattern
% >@@> primates_mean = mean(primate_ds.samples, 1); bugs_mean = mean(bug_ds.samples, 1); primates_minus_bugs = primates_mean - bugs_mean; % <@@<
Store and visualize the results
Finally save the result as a dataset with the original header. Just replace ds.samples with the result and remove the sample attributes. Then convert back to nifti and save it using the cosmo_map2fmri function.
% >@@> ds_primates_minus_bugs = ds; % make a copy ds_primates_minus_bugs.samples = primates_minus_bugs; ds_primates_minus_bugs.sa = struct(); cosmo_check_dataset(ds_primates_minus_bugs); % good practice % store to disc output_path = config.output_data_path; output_fn = fullfile(output_path, 'primates_minus_bugs.nii'); ni = cosmo_map2fmri(ds_primates_minus_bugs, output_fn); % <@@<
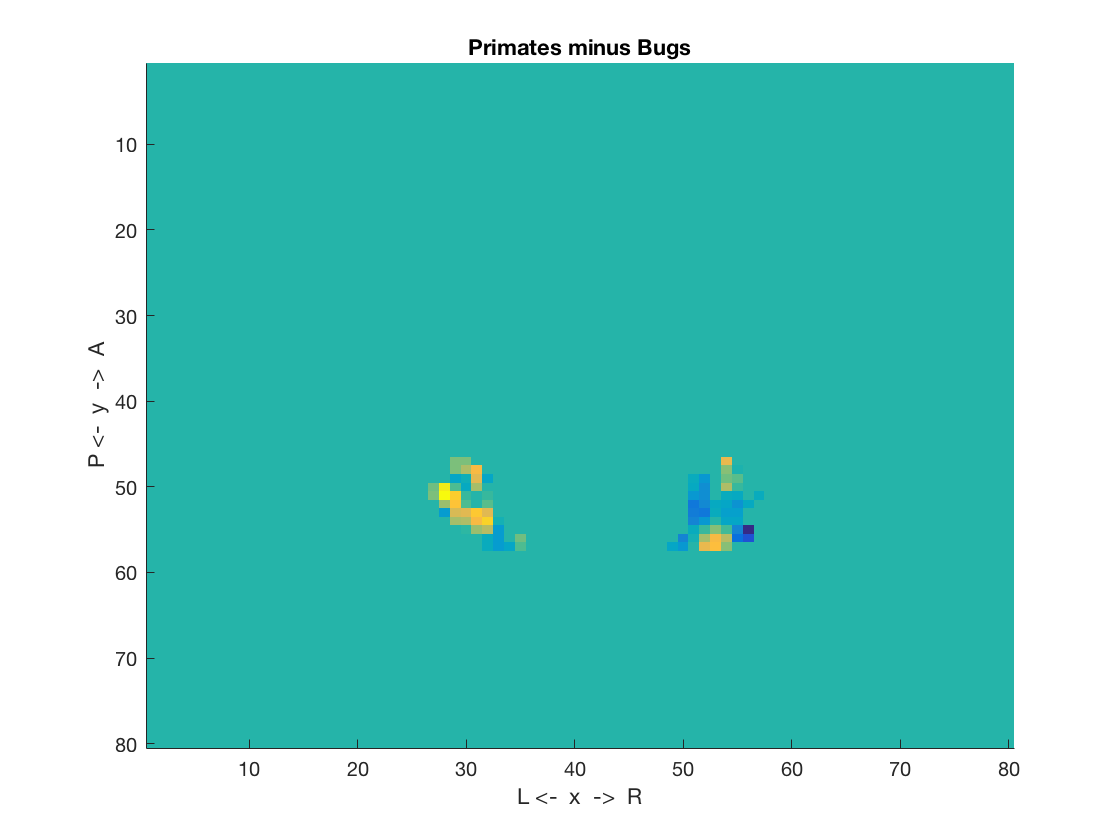
Plot results
figure; % ... using cosmo_plot_slices % >@@> cosmo_plot_slices(ds_primates_minus_bugs); % <@@< % ... using AFNI, FSL, or Matlab's imagesc figure; % >@@> imagesc(rot90(ni.img(:, :, 4))); title('Primates minus Bugs'); box off; ylabel('P <- y -> A'); xlabel('L <- x -> R'); % <@@<