Searchlight for representational similarity analysis
Using cosmo_searchlight, run cross-validation with nearest neighbor classifier
- For CoSMoMVPA's copyright information and license terms, #
- see the COPYING file distributed with CoSMoMVPA. #
Contents
Define data
config = cosmo_config(); study_path = fullfile(config.tutorial_data_path, 'ak6'); data_path = fullfile(study_path, 's01'); data_fn = fullfile(data_path, 'glm_T_stats_perrun.nii'); mask_fn = fullfile(data_path, 'brain_mask.nii'); targets = repmat(1:6, 1, 10)'; ds = cosmo_fmri_dataset(data_fn, ... 'mask', mask_fn, ... 'targets', targets); % compute average for each unique target, so that the dataset has 6 % samples - one for each target ds = cosmo_fx(ds, @(x)mean(x, 1), 'targets', 1); models_path = fullfile(study_path, 'models'); load(fullfile(models_path, 'behav_sim.mat')); load(fullfile(models_path, 'v1_model.mat'));
Set measure
Set the 'measure' and 'measure_args' to use the @cosmo_target_dsm_corr_measure measure and set its parameters to so that the target_dsm is based on behav_sim.mat
% >@@> measure = @cosmo_target_dsm_corr_measure; measure_args = struct(); measure_args.target_dsm = behav; % <@@< % Enable centering the data measure_args.center_data = true;
Run searchlight
use spherical neighborhood of 100 voxels
voxel_count = 100; % define a neighborhood using cosmo_spherical_neighborhood % >@@> nbrhood = cosmo_spherical_neighborhood(ds, 'count', voxel_count); % <@@< % Run the searchlight % >@@> results = cosmo_searchlight(ds, nbrhood, measure, measure_args); % <@@< % Save the results to disc using the following command: output_path = config.output_data_path; cosmo_map2fmri(results, ... fullfile(output_path, 'rsm_searchlight_behav.nii'));
+00:00:01 [####################] -00:00:00 mean size 99.8 +00:00:02 [####################] -00:00:00
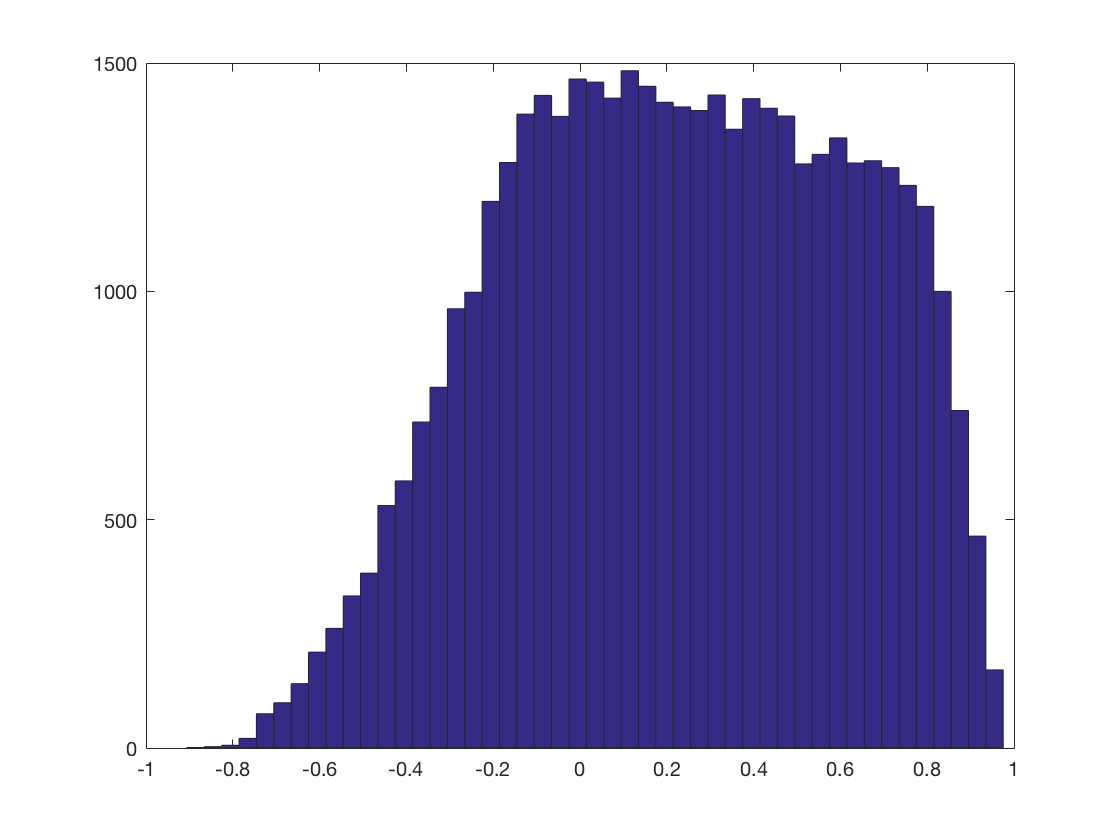
Make a histogram of correlations
hist(results.samples, 47);
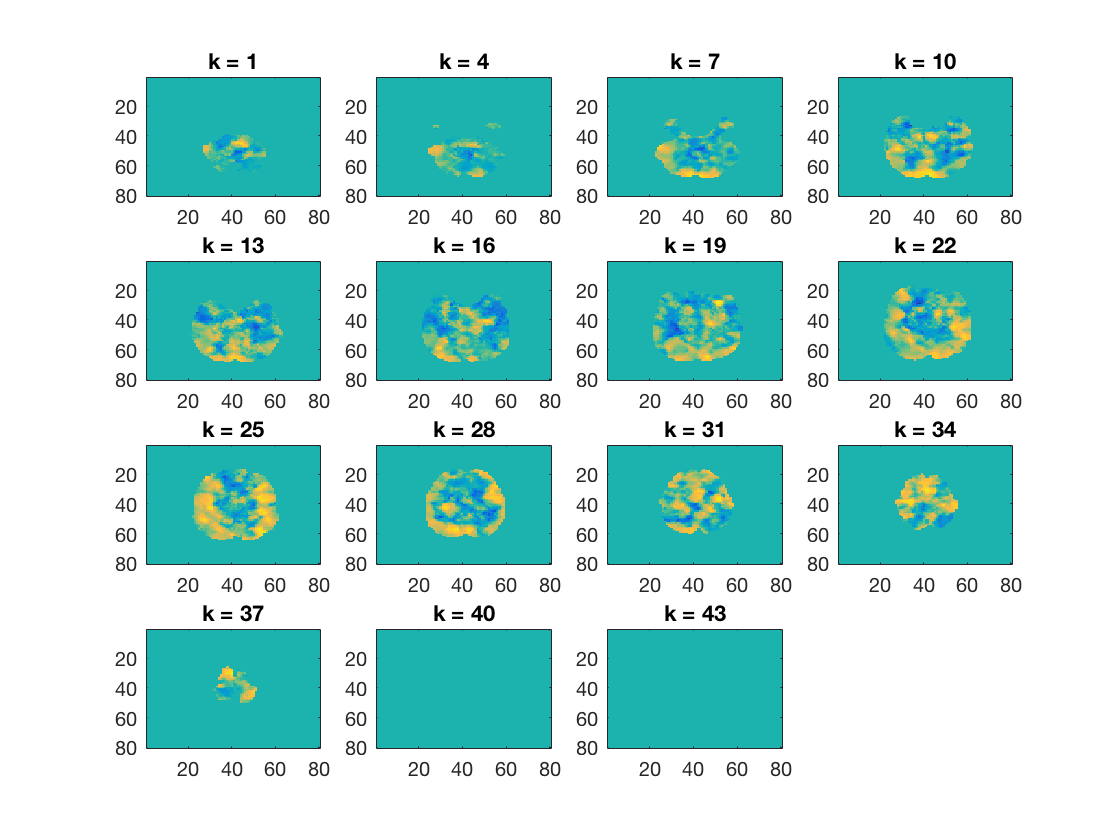
Show some slices
cosmo_plot_slices(results);
Advanced exercise: regresion-based RSA
% Using @cosmo_target_dsm_corr_measure, investigate the relative % contributions of the v1-model and behavioural similarity matrix. % % Thus, set the 'measure' and 'measure_args' to use the % @cosmo_target_dsm_corr_measure measure and set its parameters % so that the 'glm_dsm' option uses the 'behav' and 'v1_model' targets % >@@> measure = @cosmo_target_dsm_corr_measure; measure_args = struct(); measure_args.glm_dsm = {behav, v1_model}; % <@@< % Enable centering the data measure_args.center_data = true;
Run searchlight
use spherical neighborhood of 100 voxels
voxel_count = 100; % define a neighborhood using cosmo_spherical_neighborhood % >@@> nbrhood = cosmo_spherical_neighborhood(ds, 'count', voxel_count); % <@@< % Run the searchlight % >@@> glm_dsm_results = cosmo_searchlight(ds, nbrhood, measure, measure_args); % <@@< % Save the results to disc using the following command: output_path = config.output_data_path; cosmo_map2fmri(glm_dsm_results, ... fullfile(output_path, 'rsm_searchlight_glm_behav-v1.nii'));
+00:00:01 [####################] -00:00:00 mean size 99.8 +00:00:03 [####################] -00:00:00
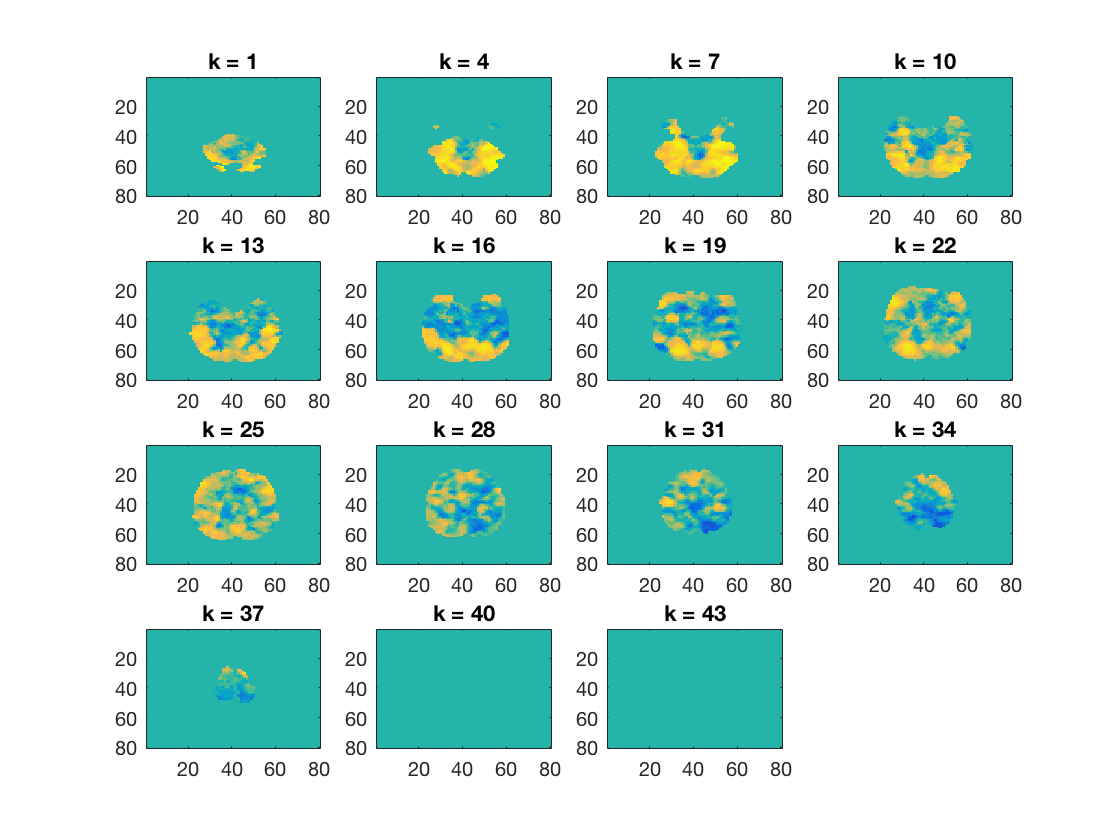
Show behavioural searchlight map
figure(); cosmo_plot_slices(cosmo_slice(glm_dsm_results, 1));
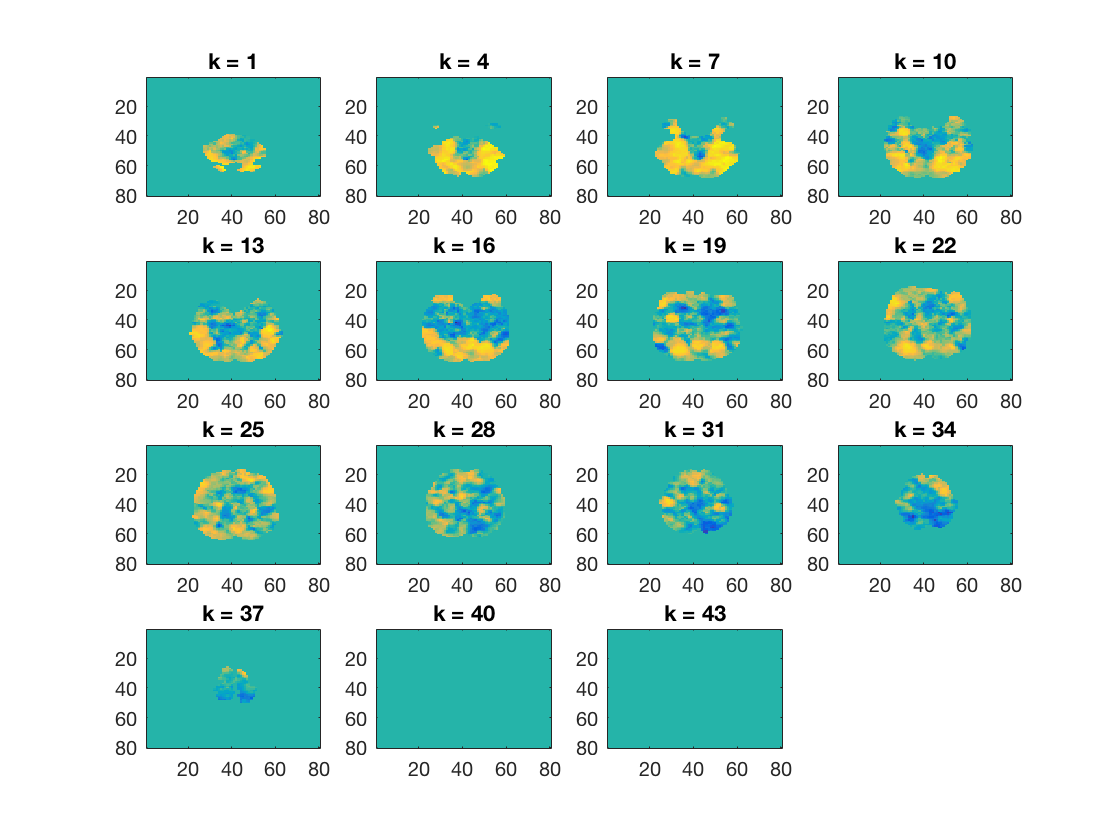
Show V1 searchlight map
figure(); cosmo_plot_slices(cosmo_slice(glm_dsm_results, 2));