Volumetric fMRI Searchlight
- For CoSMoMVPA's copyright information and license terms, #
- see the COPYING file distributed with CoSMoMVPA. #
Contents
Load data
config = cosmo_config();
data_path = fullfile(config.tutorial_data_path, 'ak6', 's01');
output_data_path = config.output_data_path;
half1_fn = fullfile(data_path, 'glm_T_stats_odd.nii');
half2_fn = fullfile(data_path, 'glm_T_stats_even.nii');
mask_fn = fullfile(data_path, 'brain_mask.nii');
half1_ds = cosmo_fmri_dataset(half1_fn, 'mask', mask_fn, ...
'targets', (1:6)', ...
'chunks', repmat(1, 6, 1));
half2_ds = cosmo_fmri_dataset(half2_fn, 'mask', mask_fn, ...
'targets', (1:6)', ...
'chunks', repmat(2, 6, 1));
ds = cosmo_stack({half1_ds, half2_ds});
ds = cosmo_remove_useless_data(ds);
Define spherical neighborhood for each feature (voxel)
radius = 3;
nbrhood = cosmo_spherical_neighborhood(ds, 'radius', radius);
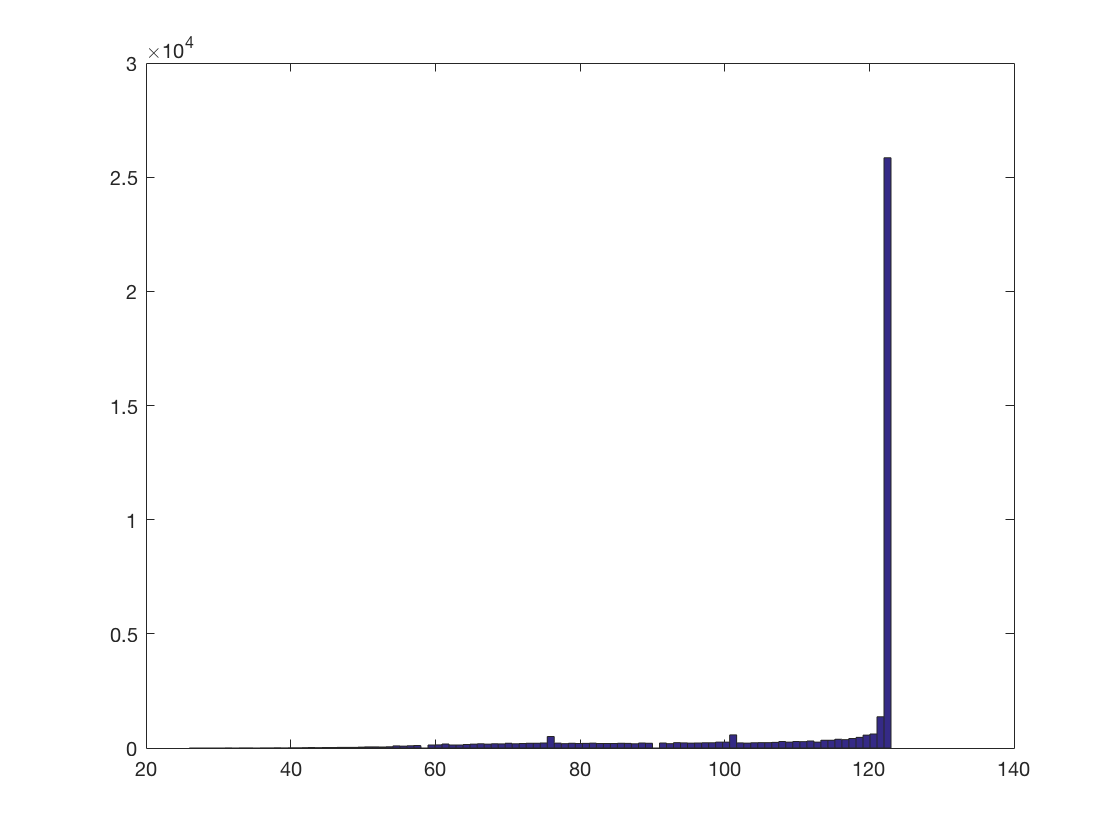
roi_sizes = cellfun(@numel, nbrhood.neighbors);
hist(roi_sizes, 100);
+00:00:00 [####################] -00:00:00 mean size 111.5
Run a searchlight with the cosmo_correlation_measure
ds_corr = cosmo_searchlight(ds, nbrhood, @cosmo_correlation_measure);
+00:00:01 [####################] -00:00:00
Visualize and store the results in a NIFTI file
cosmo_plot_slices(ds_corr);
output_fn = fullfile(output_data_path, ...
sprintf('splithalf_correlation_searchlight_r%.0f.nii', radius));
cosmo_map2fmri(ds_corr, output_fn);


