Introduction¶
Overview of the workshop¶
After an introductory presentation, it starts with basic operations of reading, writing, selecting, and aggregating dataset structures. This is followed by MVPA correlation and classficiation analysis of fMRI data in a region of interest. Subsequently, this is extended to exploratory searchlight analysis, representational similarity analysis, MEEG analysis in the space and time dimensions, and surface-based searchlights . Finally approaches to multiple comparison are discussed.
Format¶
In this workshop, all material is present on the website. Each exercise part of the workshop has three parts:
short presentation and introduction to exercise
time to work on the exercise
presentation of a possible solution to the exercise
Exercises are provided in the form of code skeletons, with part of the code left out as an exercise. Full solutions for all exercises are provided on the website.
Prerequisites¶
Matlab / Octave advanced beginner level. experience.
fMRI and/or MEEG advanced beginner level analysis experience.
Working FieldTrip installation (required for MEEG analysis).
MRI data viewer, such as MRIcron (strongly recommended).
It is recommended, prior to the course, to:
read the CoSMoMVPA manuscript (doi: 10.1101/047118, citation [OCH16]).
have the most recent CoSMoMVPA code (see Download instructions).
have a recent version of the tutorial data.
have set paths properly in
.cosmomvpa.cfg(described here)have tested that you can load and save data from and to the paths in
.cosmomvpa.cfg.
Goals of this course¶
Learn typical MVPA approaches (correlation analysis, classification analysis, representational similarity analysis).
Learn how these approaches can be applied to both fMRI and MEEG data.
- Learn how to use CoSMoMVPA to perform these analyses:
Understand the dataset structure to represent both the data itself (e.g. raw measurements or summary statistics) and its attributes (e.g. labels of conditions (targets), data acquisition run (chunks).
See how parts of the data can be selected using slicing and splitting, and combined using stacking.
Introduce measures that compute summaries of the data (such as correlation differences, classification accuracies, similarity to an a prior defined representational similarity matrix) that can be applied to both a single ROI or in a searchlight.
Learn multiple-comparison approaches.
Make yourself an independent user, so that you can apply the techniques learnt here to your own datasets.
Not covered in this course¶
Preprocessing of fMRI / MEEG data
Learning to use Matlab / Octave
Dataset types other than volumetric and surface-based fMRI data and MEEG time-locked data. (Not covered: source-space MEEG)
How to become a CoSMoMVPA developer
Datasets¶
For most of the course we will be using the AK6 dataset and the MEG obj6 dataset (described below). Although these can be downloaded separately, it is recommended however to use the full tutorial dataset.
Download link: tutorial data: datadb-v0.3.zip.
AK6 dataset¶
This dataset is used for exercises shown on the website (with answers), and you can use it to learn MVPA. It contains preprocessed data for 8 subjects from [CGG+12]. In this experiment, participants were presented with categories of six animals: 2 primates: monkeys and lemurs; 2 birds: mallard ducks and yellow-throated warblers; and 2 bugs: ladybugs and luna moths.
Download link: tutorial data: datadb-ak6-v0.3.zip.
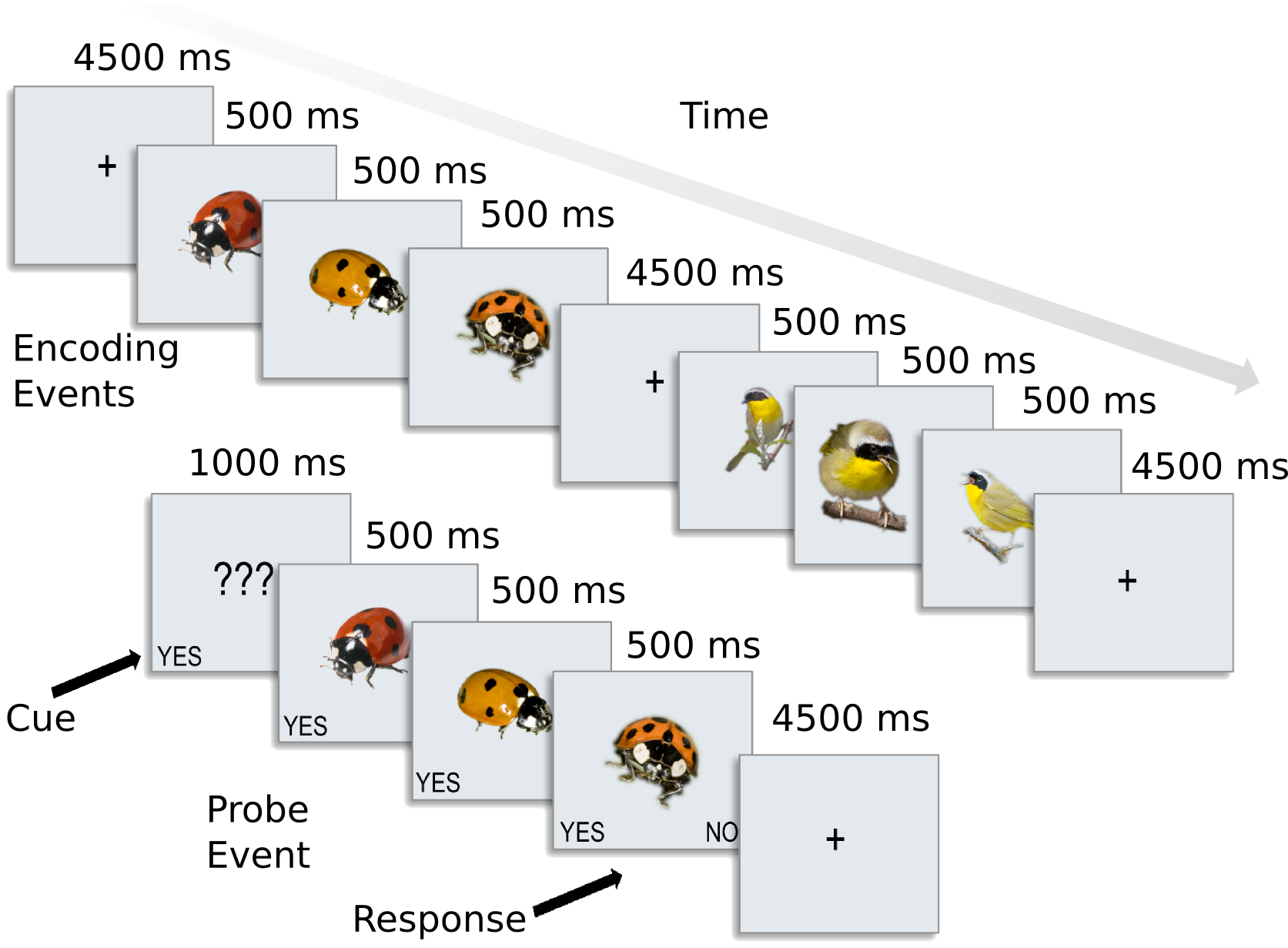
For each participant, the following data is present in the ak6 (for Animal Kingdom, 6 species) directory:
- s0[1-8]/ This directory contains fMRI data from 8 of the 12
participants studied in the experiment reported in
Connolly et al. 2012 (Code-named 'AK6' for animal
kingdom, 6-species). Each subject's subdirectory
contains the following data:
- glm_T_stats_perrun.nii A 60-volume file of EPI-data preprocessed using
AFNI up to and including fitting a general linear
model using 3dDeconvolve. Each volume contains the
t-statistics for the estimated response to a one
of the 6 stimulus categories. These estimates were
calculated independently for each of the 10 runs
in the experiment.
- glm_T_stats_even.nii Data derived from glm_T_stats_perrun.nii.
- glm_T_stats_odd.nii Each is a 6-volume file with the T-values averaged
across even and odd runs for each category.
- brain.nii Skull-stripped T1-weighted anatomical brain image.
- brain_mask.nii Whole-brain mask in EPI-space/resolution.
- vt_mask.nii Bilateral ventral temporal cortex mask similar to
that used in Connolly et al. 2012.
- ev_mask.nii Bilateral early visual cortex mask.
Also present are model similarity structures, which you can see here:
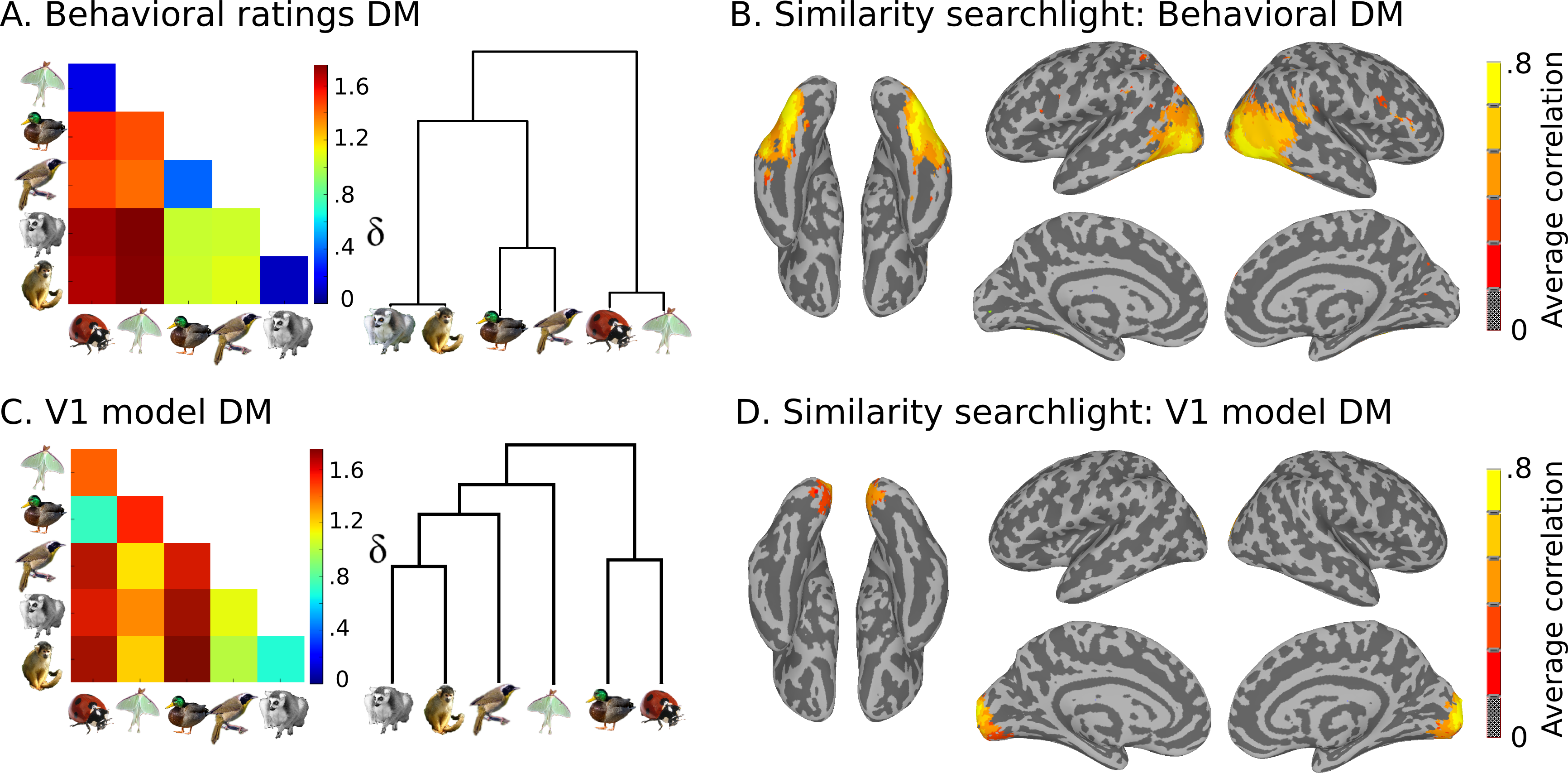
This data is stored in the models directory:
- models
- behav_sim.mat Matlab file with behavioural similarity ratings.
- v1_model.mat Matlab file with similarity values based on
low-level visual properties of the stimuli.
MEG obj6 dataset¶
This dataset is used for both the tutorial and for the assignments.
It contains MEG data from a single participant viewing images of six categories; for details see the README file.
Download link: tutorial data: datadb-meg_obj6-v0.3.zip.
Tentative schedule¶
When:
22.07.2019 - 26.07.2019.
Where:
Magdeburg, Germany; Universitätsplatz campus, Gebäude 28, room 27
For dinner and other information, see: https://www.noesseltlab.org/events-presentations/3rd-modelling-symposium-1/
Date and time |
Description |
|---|---|
Monday |
|
09:00 |
General introduction presentation |
10:30 |
Coffee break |
11:00 |
Getting started. Get Started; Dataset basics |
12:30 |
Lunch break |
14:00 |
|
15:30 |
Coffee break |
16:00-17:30 |
Split-half correlations. Split-half correlation-based MVPA with group analysis |
17:40-18:30 |
Optional: discuss your data models |
Tuesday |
|
09:00 |
Classification analysis, Classification analysis with cross-validation. |
10:30 |
Coffee break |
11:00 |
|
12:30 |
Lunch break |
14:00 |
Double dipping, Using CoSMoMVPA neighborhoods for regions of interest first part |
15:30 |
Coffee break |
16:00-17:30 |
Neighborhoods and searchlight basics. Using CoSMoMVPA neighborhoods for regions of interest |
17:40-18:30 |
Optional: discuss your data models |
Wednesday |
Free day |
Thursday |
|
09:00 |
Whole-brain fMRI searchlight. Use the searchlight with a neighborhood and a measure |
10:30 |
Coffee break |
11:00 |
M/EEG searchlight part 1: General MEEG analysis toolboxes |
12:30 |
Lunch break |
14:00 |
M/EEG searchlight part 2: General MEEG analysis toolboxes |
15:30 |
Coffee break |
16:00-17:30 |
M/EEG time generalization: MEEG time generalization |
17:40-18:30 |
Optional: discuss your data models |
Friday |
|
09:00 |
Present your data |
10:30 |
Coffee break |
11:00 |
Representational similarity analysis Representational similarity analysis |
12:30 |
Lunch break |
14:00 |
Surface-based searchlight. Surface-based fMRI searchlight |
15:30 |
Coffee break |
16:00-17:30 |
Multiple comparison correction. Concluding remarks. Using CoSMoMVPA multiple-comparison correction |
Acknowledgements¶
Thanks to Felix Ball, Emanuele Porcu, Peter Vavra, Nico Marek, Camila Agostino, Tömme Noesselt for organizing the symposium.
Contact¶
Please send an email to a@b, b=gmail.com, a=n.n.oosterhof.

